Genetic optimization of deep CNN based transfer learning for COVID-19 identification from radiography chest X-Rays
A. Environment
- Google Colaboratory: This project use Google Colaboratory for compute purpose.
- RAM: 12 GB, Disk Space: 80 GB
- Compute Engine: GPU
- Language: Python
B. Dataset
- Covid-19 Radiography Dataset
- M. E. Chowdhury, T. Rahman, A. Khandakar, R. Mazhar, M. A. Kadir, Z. B. Mahbub, K. R. Islam, M. S. Khan, A. Iqbal, N. Al Emadi, et al., Can ai help in screening viral and covid-19 pneumonia?, IEEE Access 8 (2020) 132665–132676.
- T. Rahman, A. Khandakar, Y. Qiblawey, A. Tahir, S. Kiranyaz, S. B. A.395 Kashem, M. T. Islam, S. Al Maadeed, S. M. Zughaier, M. S. Khan, et al., Exploring the effect of image enhancement techniques on covid-19 detection using chest x-ray images, Computers in biology and medicine 132 (2021) 104319.
- Sample Data: Following is the sample of dataset with class label.
C. Experimental Data
- Pickle Data: All experimental data of this project work are stored in Pickle Data.
- Data: All training histories are stored in this link.
- Models: All best models data found during optimization and exploitation are stored here.
- TL-Models: All best models data found during training of transfer learning models are stored here.
D. Codes
- **Model: **
VGG16: VGG16 model primirily used for this project. Following figure show the structure of this CNN model.

- Transfer_Learning_with_Optimization_of_Covid_19_Image_Classification.ipynb/py: Codes on optimization process on transfer learning for covid-19 identification.
collect_Data()- This function used to collect data based on the classes (folders, each folder represent a class in dataset folder) from given drive path.
- Pickle
- Pickle is used for store and retrive data from given drive path.
- `train_test_split(…)
- Used for data spliting for cross validation during training.
display_multiple_img(...)- Used for display some random data images with thier classes.
train(...), compile(...), fit(...), ...etc- Used for training the model and performance measures.
initial_individuals(model)- Used for initialization of unique individual for the initial population.
mutate(individual, model)- Used for mutation of
individualwith a mutation factor and provide unique gene.
- Used for mutation of
crossover(individuals, model)- Used for crossover operation on a population (
individuals) and provide uniqueindividualin population using uniform crossover.
- Used for crossover operation on a population (
evolve(individuals, fitness, model)- Used for evolution of
individualsbased onfitnessfor next generation. In this work evolution is done upto 30 generations.
- Used for evolution of
- Performance Measures
- After optimization of models performance measures for best 3 models are conduct based on accuracies, confusion matrix, etc.
- Exploitation_on_Transfer Learning_with_Optimization_Covid_19_Image_Classification.ipynb/py: Codes on further exploitation process on optimized transfer learning models found for covid-19 identification.
- This python file includes codes for training of optimized transfer learning models found for further 100 epochs.
- Transfer Learning_without_Optimization_Covid_19_Image_Classification.ipynb/py: Codes on straight forward transfer learning training process for covid-19 identification.
- This python file includes codes for training of straight forward transfer learning models without optimization for 300 epochs and saved best models found.
- Model construction is done randomly based on fair coin toss.
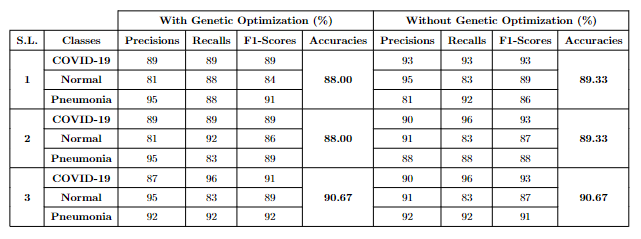
E. Results and Performance
- Results: Performance of the Transfer Learning Models with and without Genetic Optimization.
F. Dependencies
- The dataset importation from Google Drive and saving model data into Google Drive required drive permission.